Alcoholic liver disease (ALD) is a common chronic liver disease that often leads to cirrhosis. With the rising incidence, alcoholic liver disease has become the leading cause of liver transplantation, accounting for more than 50% of deaths due to liver disease. The initial stage is usually alcoholic fatty liver, which can progress to alcoholic hepatitis, liver fibrosis and cirrhosis. About 75% of alcoholic liver disease is diagnosed in the stage of cirrhosis, where liver function is severely impaired and is not suitable for drug treatment for alcohol use disorder. If alcoholic liver disease is detected early in the asymptomatic stage, intervention measures for alcohol dependence and treatment of metabolic complications can be taken to prevent or delay the progression of the disease.
On June 2, 2022, the scientific team of the Novo Nordisk Foundation Protein Research Center at the University of Copenhagen and the research group of Matthias Mann of the Max-Planck Institute in Germany published a paper entitled Noninvasive proteomic biomarkers for alcohol-related liver disease in Nature Medicine. ‘s article. reported a mass spectrometry-based proteomic assay and machine learning to construct a panel of protein biomarkers that can be used to predict liver fibrosis, hepatitis, and hepatic steatosis. The research results were listed as Research Highlights by the natural journal “Nature”.
The research team published an article in 2019 based on the detection of protein markers associated with non-alcoholic fatty liver disease (NAFLD) based on plasma proteome mass spectrometry. The analysis of the plasma proteome of 48 patients with liver cirrhosis or NAFLD found 6. 3 proteins with significant changes (ALDOB, APOM, LGALS3BP, PIGR, VTN, and AFM), of which polymeric immunoglobulin receptor (PIGR) was significantly altered in both nonalcoholic fatty liver and cirrhosis cohorts, with elevated levels, respectively 170% and 298%.
Due to the slow progression of the disease and the insidious symptoms, it brings certain challenges to the early diagnosis of alcoholic liver disease. The definitive diagnosis of hepatitis requires liver biopsy, which is difficult to accept as an invasive test, and complications occur in 1% of patients. Existing non-invasive biomarker tests have limited accuracy in the early diagnosis of alcoholic liver disease, resulting in failure to detect in time and miss the best time for intervention. Therefore, there is an urgent need for reliable diagnostic methods to screen high-risk groups such as a history of alcoholism, obesity, low physical activity, and diabetes. In addition, the molecular pathological mechanisms of alcoholic liver disease are not fully understood, and there are currently no specific interventions for the disease. Proteomic analysis of liver and plasma samples, combined with machine learning to screen molecular markers and build models, can further understand the molecular mechanism of the occurrence and development of the disease, and provide a reference for early screening and drug target discovery.
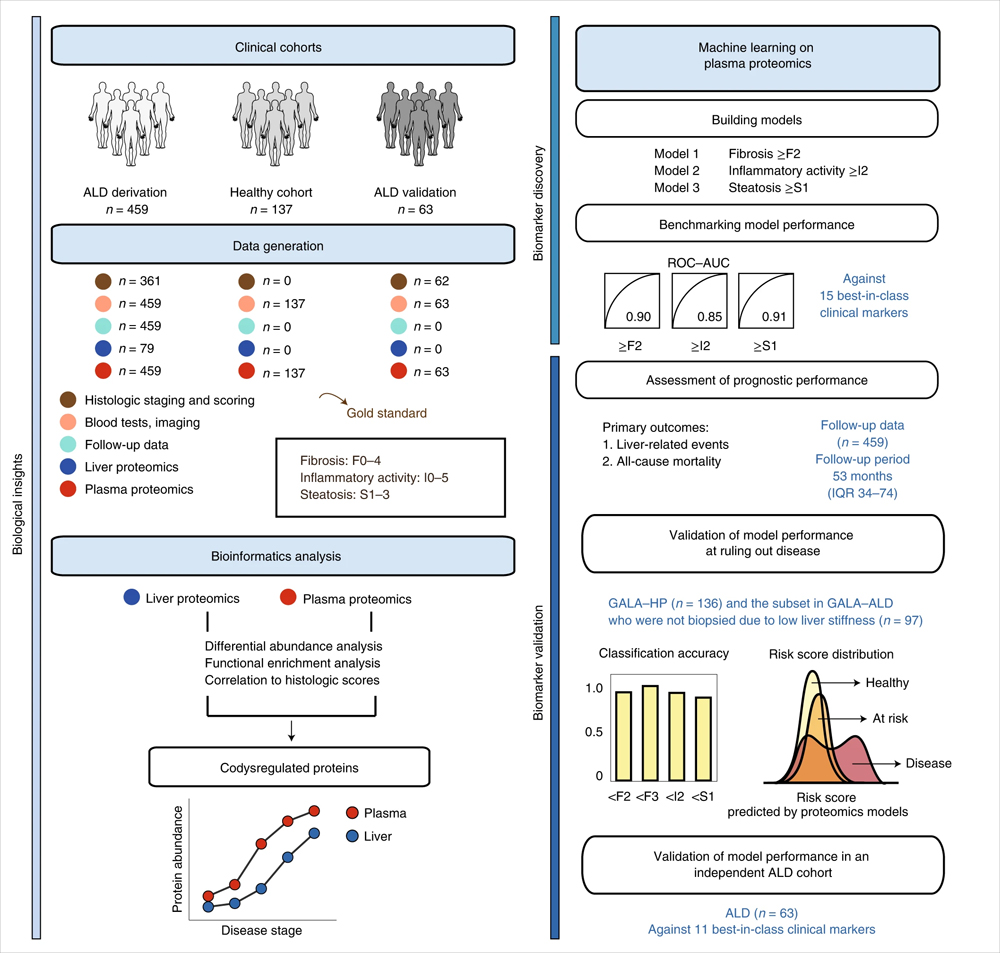
In the results of the translational study of alcoholic liver disease published this time, researchers used proteomic and machine learning models to identify a combination of biomarkers for alcoholic liver disease. Mass spectrometry-based proteomics assessed paired plasma samples and liver biopsy samples from 596 individuals (459 patients with alcoholic liver disease and 137 control individuals). The constructed machine learning model biomarker panel detects liver fibrosis, hepatitis and hepatic steatosis.
Mainly using data independent acquisition (DIA) mode, by liquid chromatography-mass spectrometry (LC-MS/MS), the plasma samples of 459 ALD patients and 137 healthy control patients were analyzed for protein enzymatic peptide fragments. Mass spectrometry analysis was performed on an Evosep One+Orbitrap Exploris 480 (Thermo Fisher Scientific).
The researchers selected 9 protein marker combinations to identify significant liver fibrosis, 6 protein marker combinations to identify mildly active hepatitis, and 12 marker combinations to identify hepatic steatosis. The three marker panels included a total of 22 unique proteins, eight of which were altered in both plasma and liver, including C7, LGALS3BP, TGFBI, ALDOB, CLEC3B, FBLN1, ATRN, and SERPINF1. The active hepatitis group had the highest proportion of co-regulatory proteins at 67% (33% and 42% in the fibrosis and steatosis groups, respectively). Among the six NAFLD candidate protein marker combinations screened in the previous stage, three proteins, ALDOB, AFM and LGALS3BP, were also detected. The mean cross-validated ROC-AUC of logistic regression models based on selected biomarker combinations were 0.90, 0.85, and 0.91, respectively, for the prediction of significant fibrosis, mildly active hepatitis, and hepatic steatosis.
Compared with existing Best-in-class (best-in-class) clinical assays, the proteomic model constructed in this study has superior performance. Validation cohorts with significant liver fibrosis and mild inflammation, further recapitulating the performance of the proteomic model, showed unique clinical advantages. For example, the F2 proteomic model excludes significant fibrosis with superior performance compared to TE or ELF blood tests routinely used to assess liver fibrosis. Accurate exclusion of disease can avoid overdiagnosis, and comprehensive diagnostic and prognostic information based on a single blood sample can reduce the number of repeated patient admissions and speed treatment decisions. The results demonstrate that plasma proteomics can guide clinical decision-making by simultaneously and accurately detecting liver fibrosis, hepatitis and steatosis in alcoholic liver disease.
Regarding the clinical translation potential of this detection technology, the researchers proposed that this technology has the advantage of achieving clinical translation through protein quantification. Compared to elastography, it has shown unique competitiveness in detecting significant fibrosis, including F2. To increase throughput and reduce costs, future targeted proteomic approaches could be developed to detect protein marker panels in a shorter time, similar to the more common and lower-cost LC-MS/MS-based vitamins currently used in clinical practice D detection. In the future, custom mass spectrometers that are easy to maintain and operate could also be developed as clinical diagnostic tools. However, the authors also point out that from the perspective of commercialization and accessibility, the possibility of developing immunological-based detection of protein marker combinations, or combining immunocapture with targeted mass spectrometry for clinical diagnostics is not excluded.
The research results provide a reference for the development of therapeutic drugs for alcoholic liver disease. For example, 20% of the up-regulated proteins are related to signaling pathways such as receptor tyrosine kinases, other potential drug targets such as G protein-coupled receptor signaling pathways. In addition, co-regulatory proteins in liver tissue and plasma, such as immune and inflammatory responses, cell adhesion, extracellular matrix (ECM), and protease inhibition, may also be potential targets for drug development.
In conclusion, this study confirms previous findings and provides potential protein targets with diagnostic, prognostic and therapeutic value in alcoholic liver disease. Biomarker combinations may be applicable to other etiologies of liver disease, such as nonalcoholic fatty liver disease with shared histological features. Future studies in larger clinical cohorts are required.